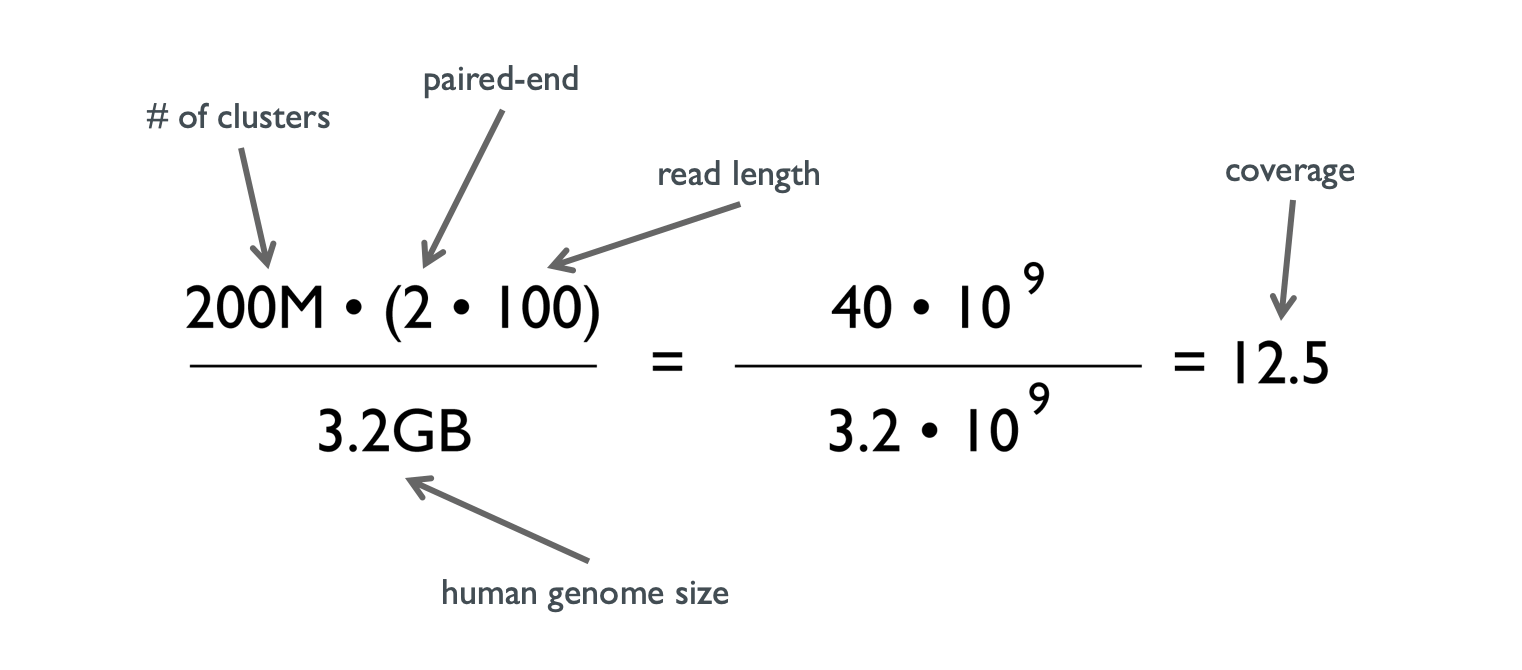
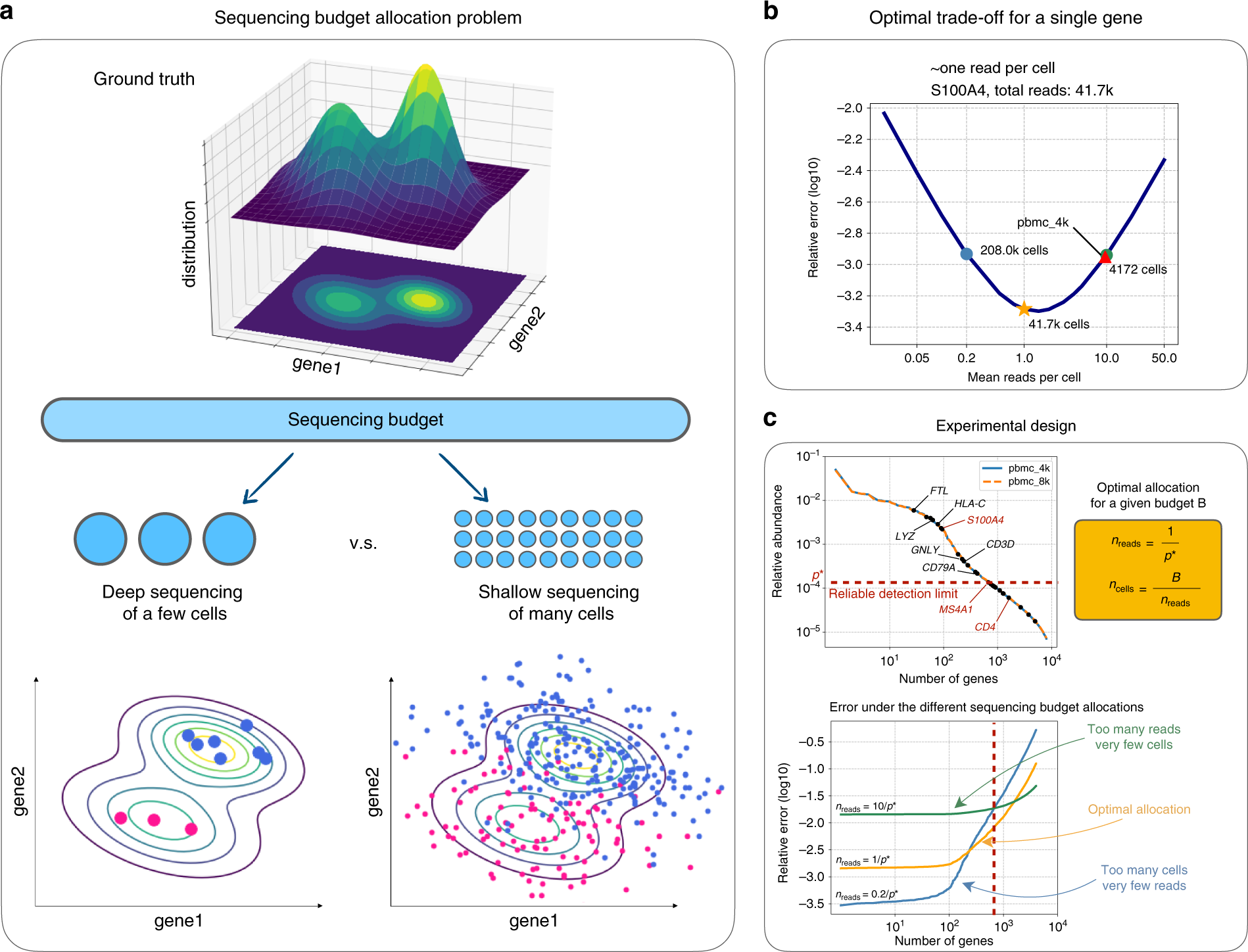
Detecting, Categorizing, and Correcting Coverage Anomalies of RNA-Seq Quantification - ScienceDirect

Detecting, Categorizing, and Correcting Coverage Anomalies of RNA-Seq Quantification - ScienceDirect
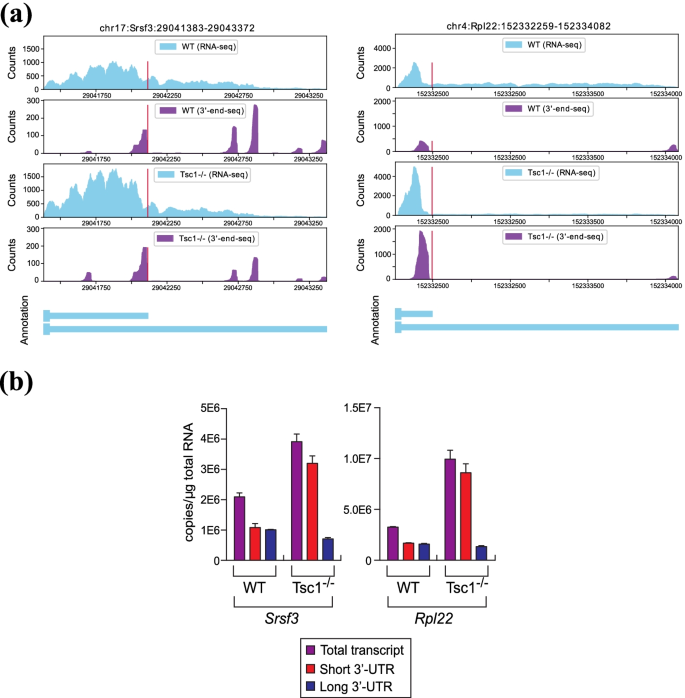
APA-Scan: detection and visualization of 3′-UTR alternative polyadenylation with RNA-seq and 3′-end-seq data | BMC Bioinformatics | Full Text
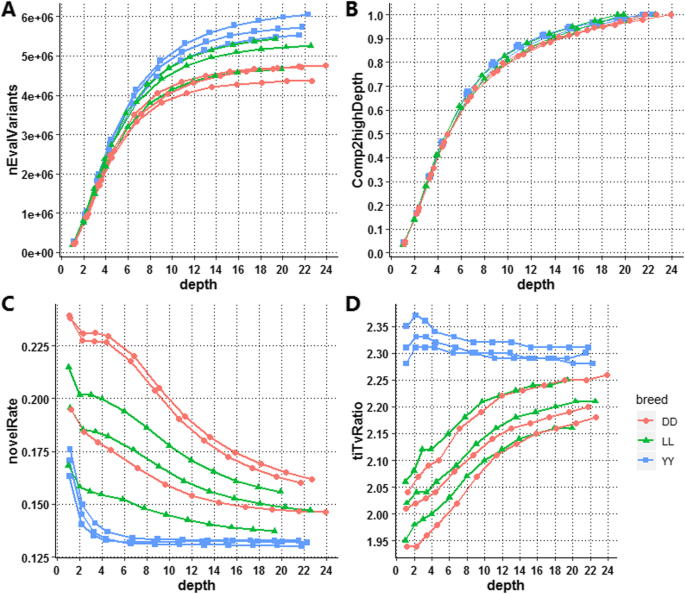
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text
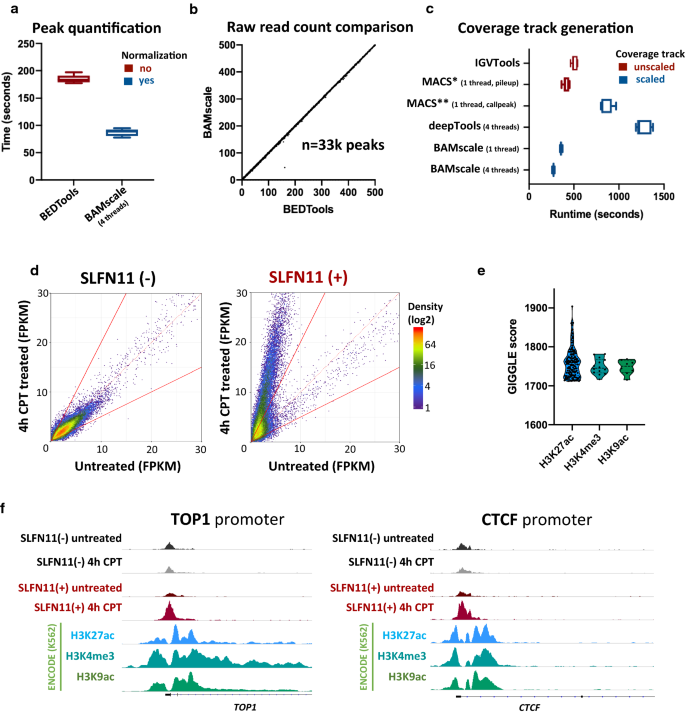
BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks | Epigenetics & Chromatin | Full Text